Section 4 Time of day analyses
In this script, we will assess if the time of day (1-hour segments) have an effect on the species richness.
4.4 What times of day have been sampled across sites?
Here we get a sense of what times of day have been visited/sampled most across sites for point count data and acoustic surveys
nSitesTime_pc <- datSubset%>%
filter(data_type == "point_count") %>%
dplyr::select(site_id, time_of_day)%>%
distinct() %>% arrange(time_of_day) %>%
count(time_of_day) %>%
mutate(data_type = "point_count")
nSitesTime_aru <- datSubset%>%
filter(data_type == "acoustic_data") %>%
dplyr::select(site_id, time_of_day)%>%
distinct() %>% arrange(time_of_day) %>%
count(time_of_day) %>%
mutate(data_type = "acoustic_data")
# This comparison lets us know that number of visits for the time_of_day - 9AM to 10AM is significantly different between acoustic data and point counts. 4.5 Estimate richness by time of day
Here we will estimate species richness for different 1-hour segments with the expectation that richness/detections would differ between point counts and acoustic surveys.
# point-count data
# estimate total abundance across all species for each site
abundance <- datSubset %>%
filter(data_type == "point_count") %>%
group_by(site_id, scientific_name,
common_name, eBird_codes, time_of_day) %>% summarise(totAbundance = sum(number)) %>%
#filter(time_of_day != "9AM to 10AM") %>%
ungroup()
# estimate richness for point count data
pc_richness_time <- abundance %>%
mutate(forRichness = case_when(totAbundance > 0 ~ 1)) %>%
group_by(site_id, time_of_day) %>%
summarise(richness = sum(forRichness)) %>%
mutate(data_type = "point_count") %>%
ungroup()
# estimate total number of detections across the acoustic data
# note: we cannot call this abundance as it refers to the total number of vocalizations across a 16-min period across all sites
detections <- datSubset %>%
filter(data_type == "acoustic_data") %>%
group_by(site_id, scientific_name,
common_name, eBird_codes, time_of_day) %>% summarise(totDetections = sum(number)) %>%
#filter(time_of_day != "9AM to 10AM") %>%
ungroup()
# estimate richness for acoustic data
aru_richness_time <- detections %>%
mutate(forRichness = case_when(totDetections > 0 ~ 1)) %>%
group_by(site_id, time_of_day) %>%
summarise(richness = sum(forRichness)) %>%
mutate(data_type = "acoustic_data") %>%
ungroup()4.6 Visualize differences in richness between point count data and acoustic data
richness_time <- bind_rows(pc_richness_time, aru_richness_time)
# visualization for different times of day
fig_richness_by_time <- richness_time %>%
grouped_ggbetweenstats(x = data_type,
y = richness,
grouping.var = time_of_day,
xlab = "Data type",
ylab = "Richness",
pairwise.display = "significant",
package = "ggsci",
palette = "default_jco",
ggplot.component = list(theme(text = element_text(family = "Century Gothic", size = 15, face = "bold"),plot.title = element_text(family = "Century Gothic",
size = 18, face = "bold"),
plot.subtitle = element_text(family = "Century Gothic",
size = 15, face = "bold",color="#1b2838"),
axis.title = element_text(family = "Century Gothic",
size = 15, face = "bold"))))
ggsave(fig_richness_by_time, filename = "figs/fig_richness_by_time.png", width = 15, height = 10, device = png(), units = "in", dpi = 300)
dev.off() 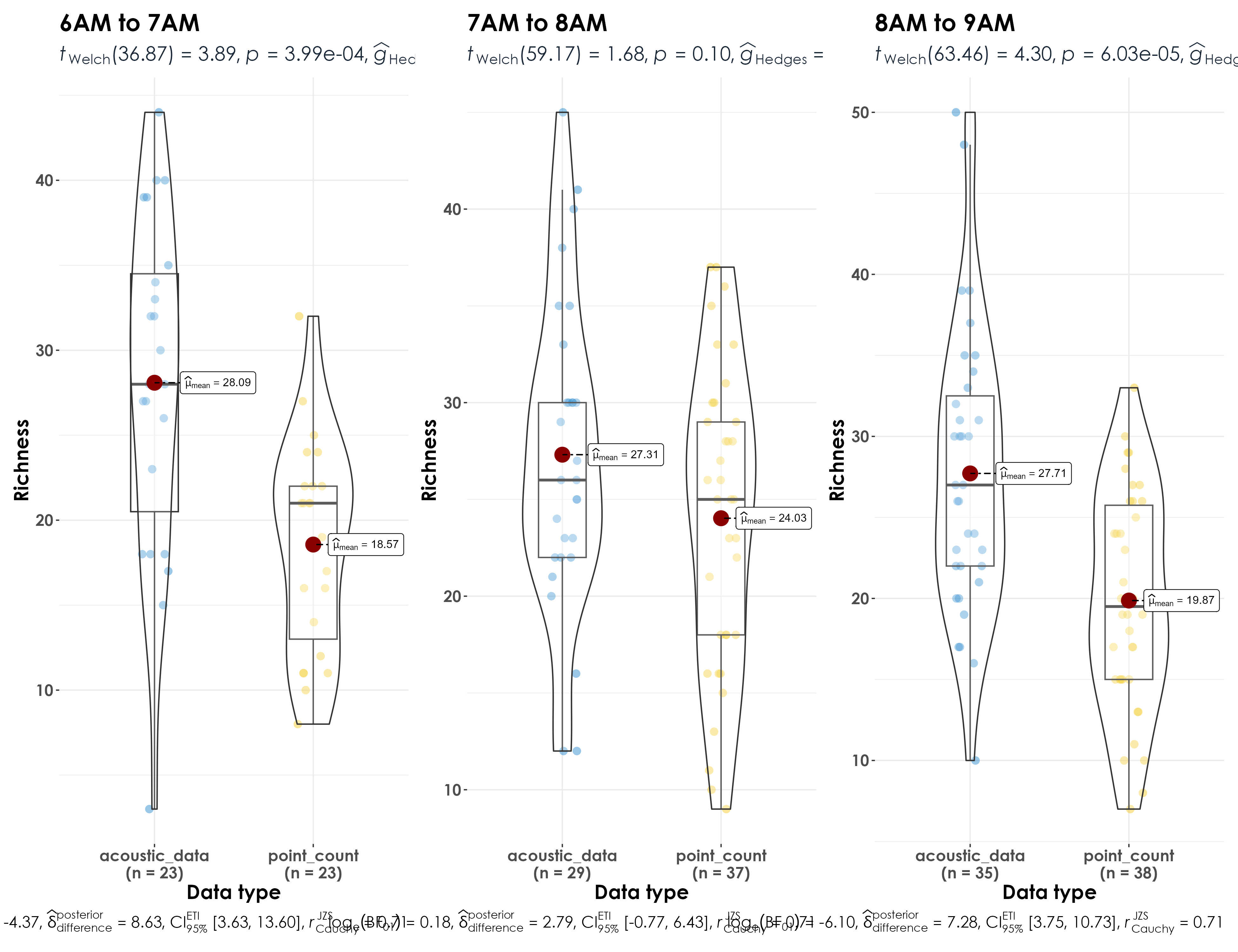
Significant differences in richness for 6am to 7am, 8am to 9am, and 9am to 10am (which can be ignored as a function of more visits in acoustic data at that time) but no differences in richness for 7am to 8am between acoustic data and point count data