Section 6 Foraging guild
Here, we ask if species of a certain foraging guild have higher vocal activity at dawn compared to dusk. Our null hypothesis/expectation is that there would be no differences in vocal activity across foraging guilds irrespective of time of day.
6.3 Filtering acoustic data to ensure sampling periods are even across dawn and dusk
Since our recording schedule was uneven (6am to 10am in the morning and 4pm to 7pm in the evening), we filter acoustic data to retain recordings between 6am and 830am and recordings made between 4pm and 630pm so that the two sampling windows capture a similar amount of time right after dawn and right before dusk.
6.4 Vocal activity across time periods
A number of factors need to be considered in further analysis: accounting for time_of_day, observed_identity for example. However, we run analyses that account for differences in calling activity by species for dawn and dusk.
# sampling effort by time_of_day
effort <- acoustic_data %>%
dplyr::select(site_id, date, time_of_day) %>%
distinct() %>%
arrange(time_of_day) %>%
count(time_of_day) %>%
rename(., nVisits = n)
# Above, we note that we had sampled ~145 site-date combinations at dawn, while ~230 site-date combinations were sampled at dusk
# total number of acoustic detections summarized across every 10-s audio file
# here, we estimate % detections at dawn and dusk, while accounting for sampling effort
vocal_act <- acoustic_data %>%
group_by(time_of_day, eBird_codes) %>%
summarise(detections = sum(number)) %>%
left_join(., species_codes[, c(1, 2, 5)],
by = "eBird_codes"
) %>%
group_by(eBird_codes) %>%
mutate(total_detections = sum(detections)) %>%
mutate(percent_detections = (detections / total_detections) * 100) %>%
ungroup()
## accouting for sampling effort and normalizing data
vocal_act <- vocal_act %>%
left_join(., effort, by = "time_of_day") %>%
mutate(normalized_detections = detections / nVisits) %>%
group_by(eBird_codes) %>%
mutate(total_normalized_detections = sum(normalized_detections)) %>%
mutate(percent_normalized_detections = (normalized_detections / total_normalized_detections) * 100) %>%
ungroup() %>%
# in our case, we have 4 species which have 100% detections in dawn, Indian blackbird, Little spiderhunter, Oriental-Magpie Robin and Purple sunbird. For these, we add a additional row specifying no detections in dusk.
add_row(
time_of_day = "dusk", eBird_codes = "pursun4", detections = 0, scientific_name = "Cinnyris asiaticus", common_name = "Purple Sunbird", total_detections = 96, percent_detections = 0, normalized_detections = 0,
percent_normalized_detections = 0, nVisits = 230, total_normalized_detections = 0.6620690
) %>%
add_row(
time_of_day = "dusk", eBird_codes = "eurbla2", detections = 0, scientific_name = "Turdus simillimus", common_name = "Indian Blackbird", total_detections = 179, percent_detections = 0, normalized_detections = 0,
percent_normalized_detections = 0, nVisits = 230,
total_normalized_detections = 1.2344828
) %>%
add_row(
time_of_day = "dusk", eBird_codes = "litspi1", detections = 0, scientific_name = "Arachnothera longirostra", common_name = "Little Spiderhunter", total_detections = 204, percent_detections = 0,
normalized_detections = 0, nVisits = 230,
percent_normalized_detections = 0,
total_normalized_detections = 1.4068966
) %>%
add_row(
time_of_day = "dusk", eBird_codes = "magrob", detections = 0, scientific_name = "Copsychus saularis", common_name = "Oriental Magpie-Robin",
total_detections = 119, percent_detections = 0,
normalized_detections = 0, nVisits = 230,
percent_normalized_detections = 0,
total_normalized_detections = 0.6620690
)6.5 Join the vocal_activity data and species trait data
vocal_act <- vocal_act %>%
left_join(trait[, c(1, 2, 29)], by = c(
"scientific_name",
"common_name"
))
## remove species that are poorly represented by a particular trophic niche
## we get rid of nectarivore species, aquatic predator and granivore
vocal_act <- vocal_act %>%
filter(trophic_niche != "Aquatic predator") %>%
filter(trophic_niche != "Granivore") %>%
filter(trophic_niche != "Nectarivore") %>%
filter(trophic_niche != "Vertivore")
## We retained 62 out of 69 species after the above filters6.6 Testing the differences among foraging guild categories using Wilcoxon test
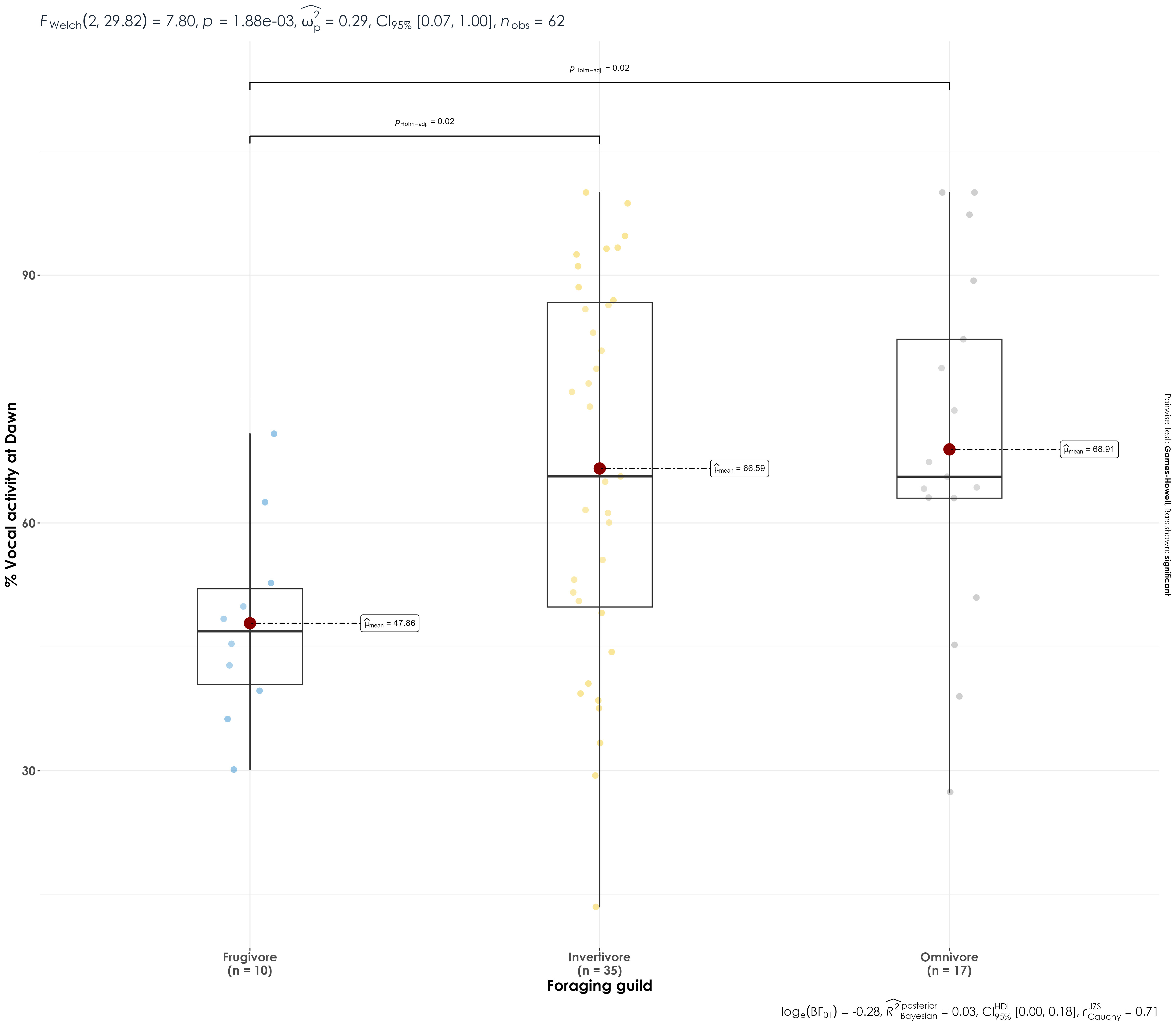
Here, we see whether there are differences in the vocal activity between invertivores, frugivores, and omnivores in dawn and dusk individually.
stat.test <- vocal_act %>%
group_by(time_of_day) %>%
wilcox_test(percent_normalized_detections ~ trophic_niche)Significant differences were observed between frugivore and omnivore species at dawn and dusk.
6.7 Comparing vocal activity at dawn between foraging guilds
fig_between_foragingGuild_vocAct <- vocal_act %>%
filter(time_of_day == "dawn") %>%
ggbetweenstats(
x = trophic_niche,
y = percent_normalized_detections,
xlab = "Foraging guild",
ylab = "% Vocal activity at Dawn",
pairwise.display = "significant",
package = "ggsci",
palette = "default_jco",
violin.args = list(width = 0),
ggplot.component = list(theme(
text = element_text(family = "Century Gothic", size = 15, face = "bold"), plot.title = element_text(
family = "Century Gothic",
size = 18, face = "bold"
),
plot.subtitle = element_text(
family = "Century Gothic",
size = 15, face = "bold", color = "#1b2838"
),
axis.title = element_text(
family = "Century Gothic",
size = 15, face = "bold"
)
))
)
ggsave(fig_between_foragingGuild_vocAct, filename = "figs/fig_percentDetections_vs_foragingGuild.png", width = 16, height = 14, device = png(), units = "in", dpi = 300)
dev.off()