Section 12 Examining changes in relative abundance over time as a function of species habitat affiliation
In this script, we will examine how relative abundance varies as a function of habitat affiliation across time periods for bird communities. We will plot species rank abundances of relative abundance to estimate how values of relative abundance has shifted for each species. In addition, we will run beta regressions to test for significance in relative abundance changes over time.
12.1 Load necessary libraries
library(dplyr)
library(stringr)
library(tidyverse)
library(scico)
library(RColorBrewer)
library(extrafont)
library(sf)
library(raster)
library(lattice)
library(data.table)
library(ggrepel)
library(report)
library(lme4)
library(glmmTMB)
library(multcomp)
library(ggstatsplot)
library(paletteer)
library(ggpubr)
library(goeveg)
library(betareg)
library(ggeffects)
library(patchwork)
library(broom)
library(ggridges)12.3 Load species trait data
## load in species trait dataset
trait_dat <- read.csv("data/species-trait-dat.csv")
# note: the classification of habitat type was created by comparing habitat affiliation data from the State of India's Birds report (v2; released in 2023).
# this classification was also vetted independently by two reviewers.
# We used the following trait classifications:
# Forest
# Grassland
# Generalist
# Our criteria for each of these classifications includes the following:
# In the 1850s, the land cover change analysis revealed that the majority of the landscape was largely grasslands and forests. However in 2018, the landscape is largely a mosaic of human-modified land cover types - including wooded habitats (mostly comprising timber plantations and degraded forests), tea plantations, followed by forests and grasslands.
# While the State of India's Birds provides a habitat classification/criteria based on contemporary use of habitats by a species, we classified species based on our resurveys and assuming that a species would either be in a forest affiliated bird species, grassland species or a generalist species in the 1850s.
# An example in this case is the black and orange flycatcher. While it is commonly found in shola forest habitat, this species is also common in degraded wooded areas, including timber plantations today. However, historically, in the 1850s (due to limited presence of timber plantations compared to 2018), we assume that this is a forest specialist bird as no other wooded habitats apart from forests existed (predominantly) in the 1850s (See 06_land-cover-classification.Rmd)
# Other examples include bird species that prefer open habitats, scrubland and dry grassland - such species have been lumped into the Grassland category as this was the closest habitat similar to the above habitat types in 1850s. Eg. Blue-tailed bee-eater, long-tailed shrike, black-winged kite.
# If a species is truly a generalist species and is not particularly occupying/specialized on any particular forested/wooded/grassland habitat, it was classified in the generalist category
## join overall relative abundance dataframe and species trait data
relAbun <- relAbun %>%
left_join(., trait_dat, by = "common_name") %>%
filter(Habitat.type != "Wetland") %>% # remove wetland birds
ungroup()
# count distinct number of species affiliated with a particular habitat type
spCount <- relAbun[, c(1, 8)] %>%
distinct() %>%
group_by(Habitat.type) %>%
count()
# Based on the above criteria, 57 species were classified as forest birds, 9 species were classified as grassland birds, 19 species were classified as generalist species.
# visualization by habitat affiliation
relAbun$Habitat.type <- factor(relAbun$Habitat.type, levels = c("Forest", "Grassland", "Generalist"))
fig_relAbun_habitat <- grouped_ggbetweenstats(
data = relAbun,
x = year,
y = relAbundance,
grouping.var = `Habitat.type`,
xlab = "Habitat.type",
ylab = "Relative Abundance",
p.adjust.method = "fdr",
ggplot.component = list(ggplot2::scale_color_discrete(c("#2c7fb8", "#025a05", "#e34a33"))),
violin.args = list(width = 0)
)
ggsave(fig_relAbun_habitat, filename = "figs/fig_relAbun_habitat_landscapeLevel.png", width = 17, height = 7, device = png(), units = "in", dpi = 300)
dev.off()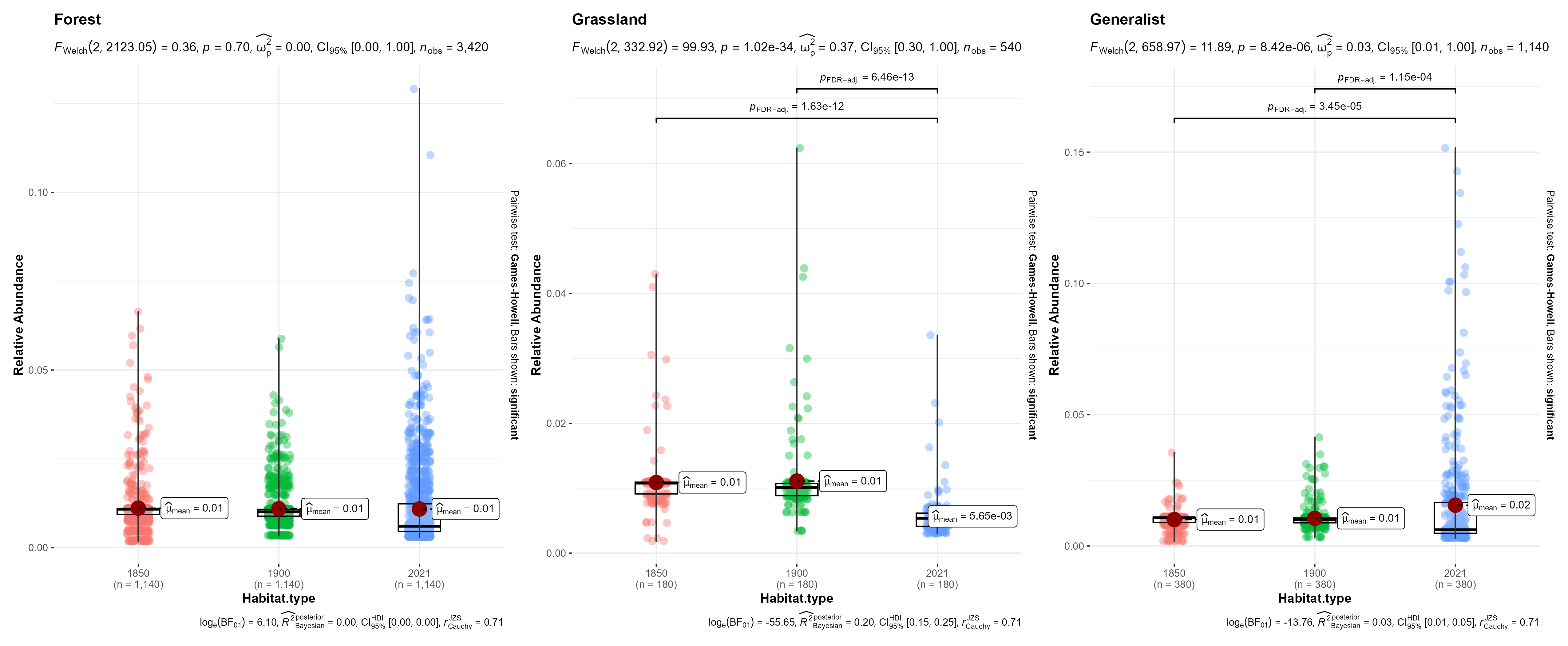
12.4 Analysis to test for significant differences in relative abundances by species across habitat types
## grassland birds
grassland_birds <- relAbun %>%
filter(Habitat.type == "Grassland")
plots <- list()
for (i in 1:length(unique(grassland_birds$common_name))) {
a <- unique(grassland_birds$common_name)[i]
# subset data for plotting
sp <- grassland_birds[grassland_birds$common_name == a, ]
p <- sp %>%
ggbetweenstats(
x = year,
y = relAbundance,
xlab = "Time Period",
ylab = "Relative Abundance",
pairwise.display = "significant",
package = "ggsci",
palette = "default_jco",
title = a,
violin.args = list(width = 0),
plotgrid.args = list(nrow = 3),
ggplot.component = list(theme(
text = element_text(family = "Century Gothic", size = 15, face = "bold"), plot.title = element_text(
family = "Century Gothic",
size = 18, face = "bold"
),
plot.subtitle = element_text(
family = "Century Gothic",
size = 15, face = "bold", color = "#1b2838"
),
axis.title = element_text(
family = "Century Gothic",
size = 15, face = "bold"
)
))
)
## save plot object
plots[[i]] <- p
}
# save as a single pdf
cairo_pdf(
filename = "figs/grassland-bird-relative-abundance-by-TimePeriod.pdf",
width = 13, height = 12,
onefile = TRUE
)
plots
dev.off()
## forest birds
forest_birds <- relAbun %>%
filter(Habitat.type == "Forest")
plots <- list()
for (i in 1:length(unique(forest_birds$common_name))) {
a <- unique(forest_birds$common_name)[i]
# subset data for plotting
sp <- forest_birds[forest_birds$common_name == a, ]
p <- sp %>%
ggbetweenstats(
x = year,
y = relAbundance,
xlab = "Time Period",
ylab = "Relative Abundance",
pairwise.display = "significant",
package = "ggsci",
palette = "default_jco",
title = a,
violin.args = list(width = 0),
plotgrid.args = list(nrow = 3),
ggplot.component = list(theme(
text = element_text(family = "Century Gothic", size = 15, face = "bold"), plot.title = element_text(
family = "Century Gothic",
size = 18, face = "bold"
),
plot.subtitle = element_text(
family = "Century Gothic",
size = 15, face = "bold", color = "#1b2838"
),
axis.title = element_text(
family = "Century Gothic",
size = 15, face = "bold"
)
))
)
## save plot object
plots[[i]] <- p
}
# save as a single pdf
cairo_pdf(
filename = "figs/forest-bird-relative-abundance-by-TimePeriod.pdf",
width = 13, height = 12,
onefile = TRUE
)
plots
dev.off()
## generalist birds
generalist_birds <- relAbun %>%
filter(Habitat.type == "Generalist")
plots <- list()
for (i in 1:length(unique(generalist_birds$common_name))) {
a <- unique(generalist_birds$common_name)[i]
# subset data for plotting
sp <- generalist_birds[generalist_birds$common_name == a, ]
p <- sp %>%
ggbetweenstats(
x = year,
y = relAbundance,
xlab = "Time Period",
ylab = "Relative Abundance",
pairwise.display = "significant",
package = "ggsci",
palette = "default_jco",
title = a,
violin.args = list(width = 0),
plotgrid.args = list(nrow = 3),
ggplot.component = list(theme(
text = element_text(family = "Century Gothic", size = 15, face = "bold"), plot.title = element_text(
family = "Century Gothic",
size = 18, face = "bold"
),
plot.subtitle = element_text(
family = "Century Gothic",
size = 15, face = "bold", color = "#1b2838"
),
axis.title = element_text(
family = "Century Gothic",
size = 15, face = "bold"
)
))
)
## save plot object
plots[[i]] <- p
}
# save as a single pdf
cairo_pdf(
filename = "figs/generalist-bird-relative-abundance-by-TimePeriod.pdf",
width = 13, height = 12,
onefile = TRUE
)
plots
dev.off()12.5 Summary of relative abundance changes by species as a function of habitat affiliation
Above, we performed an ANOVA to test if there are significant differences in species-specific means of relative abundances.
For grassland bird species (n=9) 8/9 species showed a significant difference and a decrease in relative abundance between historical and modern survey periods. No differences were observed across time periods for the Pied Bushchat
For Forest birds:(n=57) 27 bird species showed a significant decrease in relative abundance in the modern survey period when compared to either one of the historical time periods. 22 bird species showed no differences in relative abundances across time periods/remained stable. 8 bird species showed a significant increase in relative abundance in the modern survey period when compared to either historical survey periods.
~52.6% of birds have remained stable and/or increase in relative abundance over time.
For generalist birds (n=19): 10 birds showed no significant differences in relative abundance across time periods. 4 bird species showed significant increase in the modern survey period compared to either one of the historical time periods. 5 birds showed a significant decrease in relative abundance in the modern survey period compared to either one of the historical survey periods.
~73.6% of birds remained stable or increased in relative abundance over time
12.6 Has relative abundance increased or decreased for each species over time?
Based on the above plots and levels of significance for decrease in relative abundance or we create an additional column for each species to denote if a species has increased or remained stable in relative abundance over time. This column is added by manually inspecting the plots for significance and included in the species-relative-abundances.csv file.
## loading the average relative species abundances file
## this file now has a column indicating if a species increased or decreased in relative abundance
relAbun_landscape <- read.csv("results/species-relative-abundance.csv")
names(relAbun_landscape) <- c("common_name", "1850-1900", "1900-1950", "2021", "increase_decrease")
relAbun_landscape <- relAbun_landscape %>%
left_join(., trait_dat, by = "common_name") %>%
filter(Habitat.type != "Wetland") %>% # remove wetland birds
ungroup()
## merge the above file with species trait data
mean_relAbun <- relAbun %>%
dplyr::select(
common_name, historical_site_code,
year, relAbundance, Habitat.type
) %>%
group_by(common_name, year) %>%
mutate(meanRelAbun = mean(relAbundance)) %>%
dplyr::select(common_name, year, meanRelAbun) %>%
distinct() %>%
left_join(., trait_dat, by = "common_name") %>%
filter(Habitat.type != "Wetland") %>% # remove wetland birds
ungroup()
## summary pie chart
fig_relAbun_pieChart <- ggpiestats(
# arguments relevant for `ggpiestats()`
data = relAbun_landscape,
x = increase_decrease,
y = Habitat.type,
digits.perc = 0,
plotgrid.args = list(nrow = 2),
ggplot.component = list(theme(
text = element_text(family = "Century Gothic", size = 15, face = "bold"), plot.title = element_text(
family = "Century Gothic",
size = 18, face = "bold"
),
plot.subtitle = element_text(
family = "Century Gothic",
size = 15, face = "bold", color = "#1b2838"
),
axis.title = element_text(
family = "Century Gothic",
size = 15, face = "bold"
)
))
)
ggsave(fig_relAbun_pieChart, filename = "figs/fig_relativeAbundance_pieChart_increase_decrease.png", width = 15, height = 6, device = png(), units = "in", dpi = 300)
dev.off()
## Now that we have a summary pie chart showcasing the number of species that increased/remained stable and decreased in relative abundance over time, we can plot the relative abundance for the year 2021 as rank abundance plots
## add increase_decrease column
mean_relAbun <- left_join(mean_relAbun, relAbun_landscape[, c(1, 5)])
## separately for grassland, forest and generalist birds
fig_linePlot_grass <- mean_relAbun %>%
filter(Habitat.type == "Grassland") %>%
ggplot(., aes(
x = year, y = meanRelAbun, group = common_name,
color = increase_decrease
)) +
geom_line(alpha = 0.9) +
geom_point(size = 2) +
geom_label_repel(
data = . %>% filter(year == "1850"),
aes(label = common_name),
hjust = 1,
direction = "y",
nudge_x = -0.1,
size = 3,
box.padding = 0.5,
point.padding = 0.3,
segment.color = "lightgrey",
max.overlaps = Inf,
force = 10,
label.padding = 0.15,
label.r = 0.15,
fill = "white",
label.size = 0.3,
family = "Century Gothic"
) +
scale_x_discrete() +
scale_y_continuous(limits = c(0, 0.04)) +
scale_color_manual(values = c(
"decrease" = "#D95F02",
"stable/increase" = "#1B9E77"
)) +
labs(
x = "Year",
y = "Mean Relative Abundance",
title = "Change in Species Relative Abundance (1850-2021)",
color = "Trend"
) +
theme_bw() +
theme(
axis.title = element_text(
family = "Century Gothic",
size = 14, face = "bold"
),
axis.text = element_text(
family = "Century Gothic",
size = 12
),
axis.text.x = element_text(
angle = 90, vjust = 0.5,
hjust = 1
)
)
ggsave(fig_linePlot_grass, filename = "figs/fig_linePlot_grassland.svg", width = 9, height = 10, device = svg(), units = "in", dpi = 300)
dev.off()
## forest birds
fig_linePlot_forest <- mean_relAbun %>%
filter(Habitat.type == "Forest") %>%
ggplot(., aes(
x = year, y = meanRelAbun, group = common_name,
color = increase_decrease
)) +
geom_line(alpha = 0.9) +
geom_point(size = 2) +
# Labels for "decrease" (left side)
geom_label_repel(
data = . %>%
filter(
year == "1850",
increase_decrease == "decrease"
),
aes(label = common_name),
hjust = 1,
direction = "y",
nudge_x = -1.2,
size = 2.5,
box.padding = 1.2,
point.padding = 0.5,
segment.color = "lightgrey",
segment.curvature = 0,
force = 50,
min.segment.length = 0,
max.overlaps = Inf,
family = "Century Gothic",
fill = "white",
label.padding = 0.15,
label.r = 0.15,
label.size = 0.3,
ylim = c(0, 0.04)
) +
# Labels for "stable/increase" (right side)
geom_label_repel(
data = . %>%
filter(
year == "2021",
increase_decrease == "stable/increase"
),
aes(label = common_name),
hjust = 0,
direction = "y",
nudge_x = 1.2,
size = 2.5,
box.padding = 1.2,
point.padding = 0.5,
segment.color = "lightgrey",
segment.curvature = 0,
force = 50,
min.segment.length = 0,
max.overlaps = Inf,
family = "Century Gothic",
fill = "white",
label.padding = 0.15,
label.r = 0.15,
label.size = 0.3,
ylim = c(0, 0.04)
) +
scale_x_discrete(expand = expansion(mult = c(1, 1))) +
scale_y_continuous(limits = c(0, 0.04)) +
scale_color_manual(values = c(
"decrease" = "#D95F02",
"stable/increase" = "#1B9E77"
)) +
labs(
x = "Year",
y = "Mean Relative Abundance",
title = "Change in Species Relative Abundance (1850-2021)",
color = "Trend"
) +
theme_bw() +
theme(
axis.title = element_text(
family = "Century Gothic",
size = 14, face = "bold"
),
axis.text = element_text(
family = "Century Gothic",
size = 12
),
axis.text.x = element_text(
angle = 90, vjust = 0.5,
hjust = 1
)
)
ggsave(fig_linePlot_forest, filename = "figs/fig_linePlot_forest.svg", width = 15, height = 10, device = svg(), units = "in", dpi = 300)
dev.off()
## generalist birds
# please note that the y-lim was modified when compared to grassland and forest birds
fig_linePlot_generalist <- mean_relAbun %>%
filter(Habitat.type == "Generalist") %>%
ggplot(., aes(
x = year, y = meanRelAbun, group = common_name,
color = increase_decrease
)) +
geom_line(alpha = 0.9) +
geom_point(size = 2) +
# Labels for "decrease" (left side)
geom_label_repel(
data = . %>%
filter(
year == "1850",
increase_decrease == "decrease"
),
aes(label = common_name),
hjust = 1,
direction = "y",
nudge_x = -1.2,
size = 2.5,
box.padding = 1.2,
point.padding = 0.5,
segment.color = "lightgrey",
segment.curvature = 0,
force = 50,
min.segment.length = 0,
max.overlaps = Inf,
family = "Century Gothic",
fill = "white",
label.padding = 0.15,
label.r = 0.15,
label.size = 0.3,
ylim = c(0, 0.04)
) +
# Labels for "stable/increase" (right side)
geom_label_repel(
data = . %>%
filter(
year == "2021",
increase_decrease == "stable/increase"
),
aes(label = common_name),
hjust = 0,
direction = "y",
nudge_x = 1.2,
size = 2.5,
box.padding = 1.2,
point.padding = 0.5,
segment.color = "lightgrey",
segment.curvature = 0,
force = 50,
min.segment.length = 0,
max.overlaps = Inf,
family = "Century Gothic",
fill = "white",
label.padding = 0.15,
label.r = 0.15,
label.size = 0.3,
ylim = c(0, 0.04)
) +
scale_x_discrete(expand = expansion(mult = c(1, 1))) +
scale_y_continuous(limits = c(0, 0.04)) +
scale_color_manual(values = c(
"decrease" = "#D95F02",
"stable/increase" = "#1B9E77"
)) +
labs(
x = "Year",
y = "Mean Relative Abundance",
title = "Change in Species Relative Abundance (1850-2021)",
color = "Trend"
) +
theme_bw() +
theme(
axis.title = element_text(
family = "Century Gothic",
size = 14, face = "bold"
),
axis.text = element_text(
family = "Century Gothic",
size = 12
),
axis.text.x = element_text(
angle = 90, vjust = 0.5,
hjust = 1
)
)
ggsave(fig_linePlot_generalist, filename = "figs/fig_linePlot_generalist.svg", width = 12, height = 9, device = svg(), units = "in", dpi = 300)
dev.off()12.7 Box plots of relative abundance by species trait across time periods
# grassland birds
fig_boxPlot_grass <- ggplot(grassland_birds, aes(x = common_name, y = relAbundance, fill = factor(year))) +
# geom_col()+
geom_boxplot(alpha = 0.7) +
scale_fill_manual(values = c("#2c7fb8", "#025a05", "#e34a33")) +
theme_bw() +
labs(
x = "\nCommon Name",
y = "Relative Abundance\n"
) +
theme(
axis.title = element_text(
family = "Century Gothic",
size = 14, face = "bold"
),
axis.text = element_text(
family = "Century Gothic",
size = 12
),
axis.text.x = element_text(
angle = 90, vjust = 0.5,
hjust = 1
)
)
ggsave(fig_boxPlot_grass, filename = "figs/fig_boxPlot_grassland.png", width = 15, height = 10, device = png(), units = "in", dpi = 300)
dev.off()
# forest birds
fig_boxPlot_forest <- ggplot(forest_birds, aes(x = common_name, y = relAbundance, fill = factor(year))) +
geom_boxplot(alpha = 0.7) +
scale_fill_manual(values = c("#2c7fb8", "#025a05", "#e34a33")) +
theme_bw() +
labs(
x = "\nCommon Name",
y = "Relative Abundance\n"
) +
theme(
axis.title = element_text(
family = "Century Gothic",
size = 14, face = "bold"
),
axis.text = element_text(
family = "Century Gothic",
size = 12
),
axis.text.x = element_text(
angle = 90, vjust = 0.5,
hjust = 1
)
)
ggsave(fig_boxPlot_forest, filename = "figs/fig_boxPlot_forest.png", width = 22, height = 10, device = png(), units = "in", dpi = 300)
dev.off()
# generalist birds
fig_boxPlot_generalist <- ggplot(generalist_birds, aes(x = common_name, y = relAbundance, fill = factor(year))) +
geom_boxplot(alpha = 0.7) +
scale_fill_manual(values = c("#2c7fb8", "#025a05", "#e34a33")) +
theme_bw() +
labs(
x = "\nCommon Name",
y = "Relative Abundance\n"
) +
theme(
axis.title = element_text(
family = "Century Gothic",
size = 14, face = "bold"
),
axis.text = element_text(
family = "Century Gothic",
size = 12
),
axis.text.x = element_text(
angle = 90, vjust = 0.5,
hjust = 1
)
)
ggsave(fig_boxPlot_generalist, filename = "figs/fig_boxPlot_generalist.png", width = 15, height = 10, device = png(), units = "in", dpi = 300)
dev.off()
# Grassland birds are the biggest losers; generalist birds are the biggest winners while forest birds show mixed responses depending on the species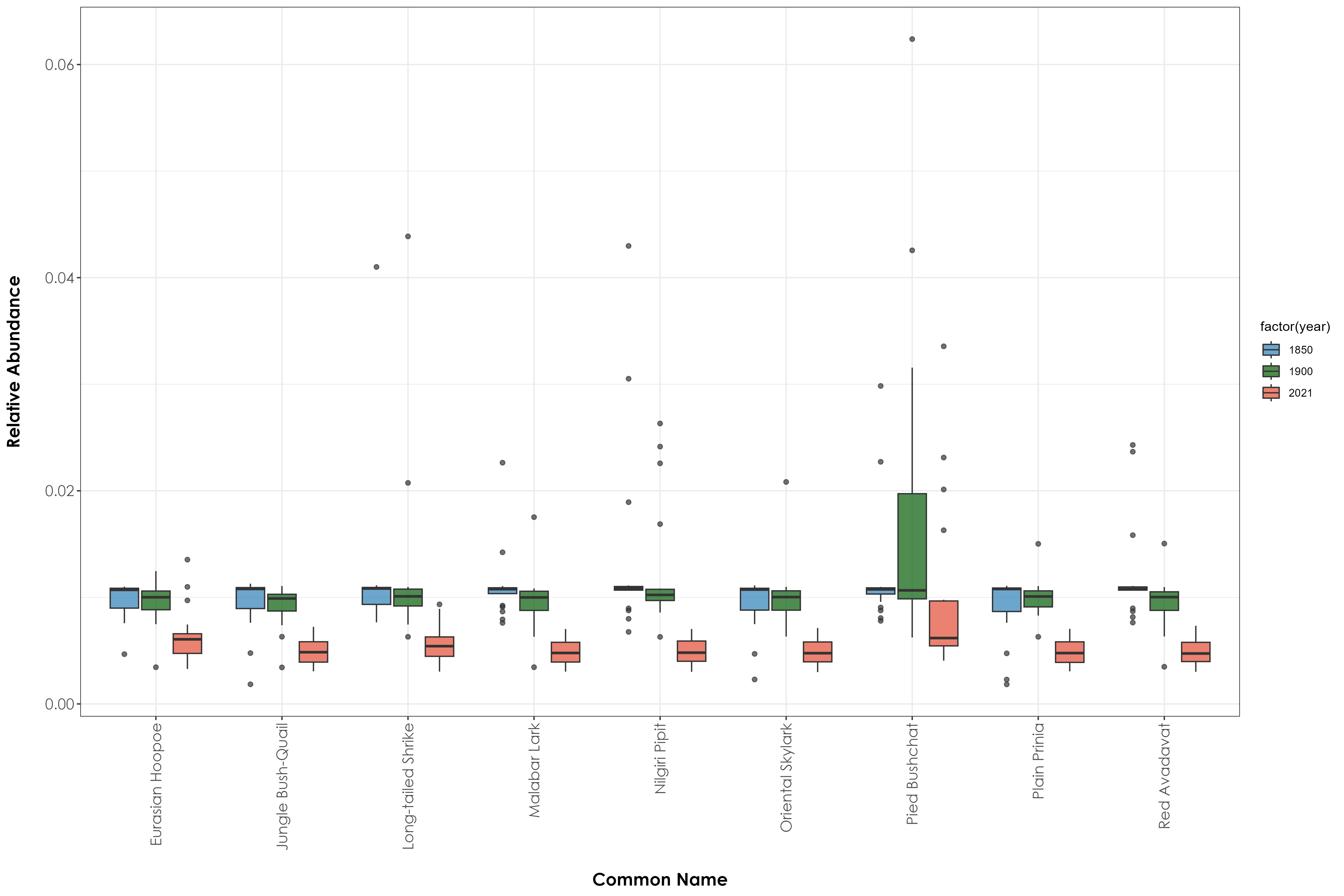
12.8 Are there significant decreases in relative abundance by time periods across habitat affiliations?
We will run beta regressions to answer the above question as the data is bounded between zero and 1.
# change the year column to factor
relAbun$year <- factor(relAbun$year)
beta_overall <- betareg(relAbundance ~ year, data = relAbun)
summary(beta_overall)
# Call:
# betareg(formula = relAbundance ~ factor(year), data = relAbun)
#
# Standardized weighted residuals 2:
# Min 1Q Median 3Q Max
# -2.7460 -0.3421 -0.0052 0.1419 4.4420
#
# Coefficients (mean model with logit link):
# Estimate Std. Error z value Pr(>|z|)
# (Intercept) -4.42983 0.01317 -336.371 <2e-16 ***
# factor(year)1900 0.01608 0.01805 0.891 0.373
# factor(year)2021 -0.22679 0.01899 -11.940 <2e-16 ***
#
# Phi coefficients (precision model with identity link):
# Estimate Std. Error z value Pr(>|z|)
# (phi) 261.911 5.357 48.89 <2e-16 ***
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#
# Type of estimator: ML (maximum likelihood)
# Log-likelihood: 1.913e+04 on 4 Df
# Pseudo R-squared: 0.05352
# Number of iterations: 22 (BFGS) + 3 (Fisher scoring)
# plot predicted model estimates
fig_beta_overall <- ggpredict(beta_overall) %>%
plot() +
set_theme(
base = theme_bw(),
theme.font = "Century Gothic",
axis.title.size = 1.2,
axis.textsize = 1,
axis.textcolor = "black"
) +
ggtitle("Predicted values of relative abundance for all data")
ggsave(fig_beta_overall, filename = "figs/fig_beta_relAbundance_overall.png", width = 9, height = 6, device = png(), units = "in", dpi = 300)
dev.off()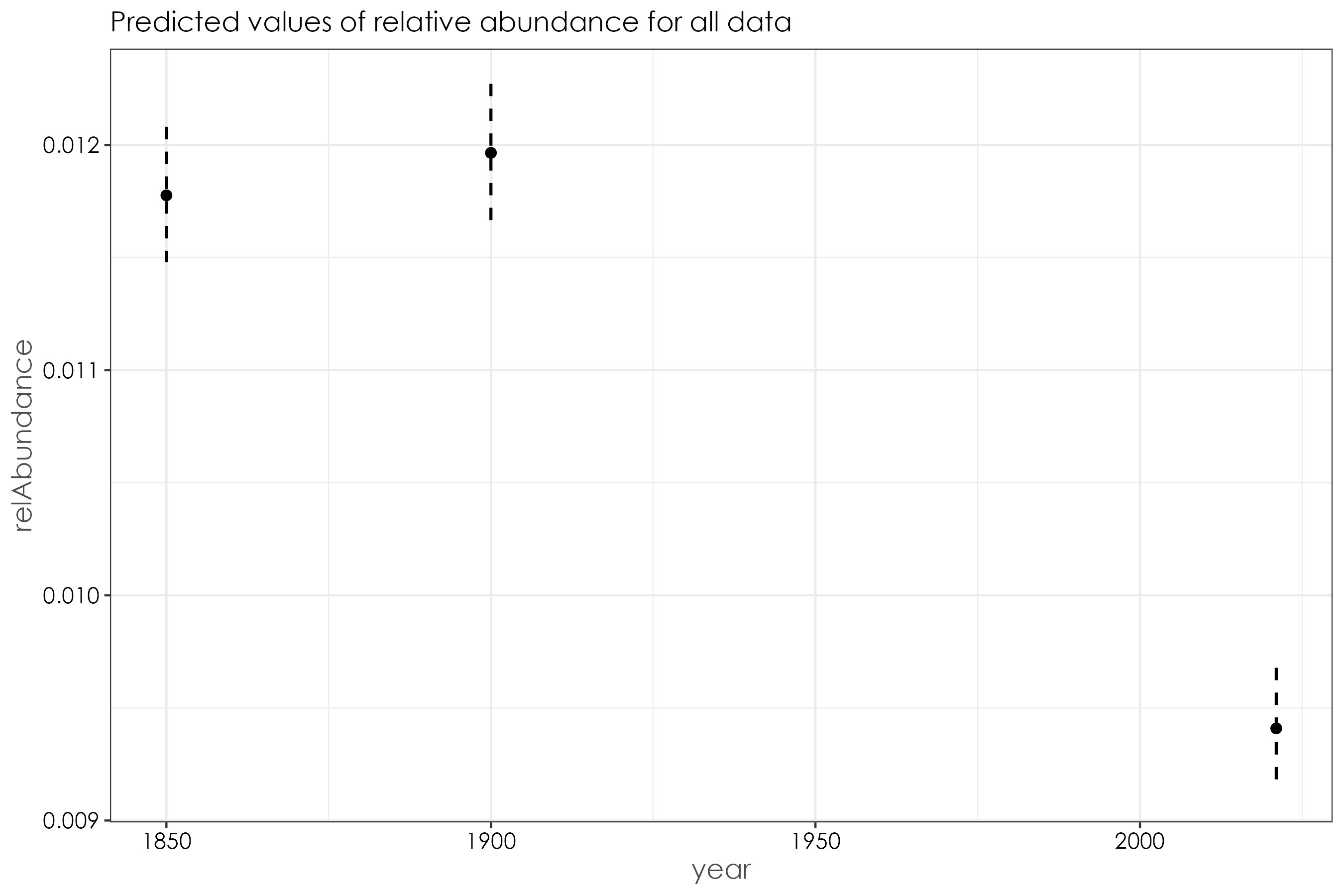
12.9 Forest birds
# change the year column to factor
forest_birds$year <- factor(forest_birds$year)
beta_forest <- betareg(relAbundance ~ year, data = forest_birds)
summary(beta_forest)
# Call:
# betareg(formula = relAbundance ~ year, data = forest_birds)
#
# Standardized weighted residuals 2:
# Min 1Q Median 3Q Max
# -2.8875 -0.3700 -0.0043 0.1296 4.3339
#
# Coefficients (mean model with logit link):
# Estimate Std. Error z value Pr(>|z|)
# (Intercept) -4.429361 0.015560 -284.660 <2e-16 ***
# year1900 0.008433 0.021403 0.394 0.694
# year2021 -0.239189 0.022568 -10.598 <2e-16 ***
#
# Phi coefficients (precision model with identity link):
# Estimate Std. Error z value Pr(>|z|)
# (phi) 281.73 7.02 40.13 <2e-16 ***
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#
# Type of estimator: ML (maximum likelihood)
# Log-likelihood: 1.294e+04 on 4 Df
# Pseudo R-squared: 0.05652
# Number of iterations: 26 (BFGS) + 2 (Fisher scoring)
# plot predicted model estimates
fig_beta_forest <- ggpredict(beta_forest) %>%
plot() +
set_theme(
base = theme_bw(),
theme.font = "Century Gothic",
axis.title.size = 1.2,
axis.textsize = 1,
axis.textcolor = "black"
) +
ggtitle("Predicted values of relative abundance for forest bird species")
ggsave(fig_beta_forest, filename = "figs/fig_beta_relAbundance_forestBirds.png", width = 9, height = 6, device = png(), units = "in", dpi = 300)
dev.off()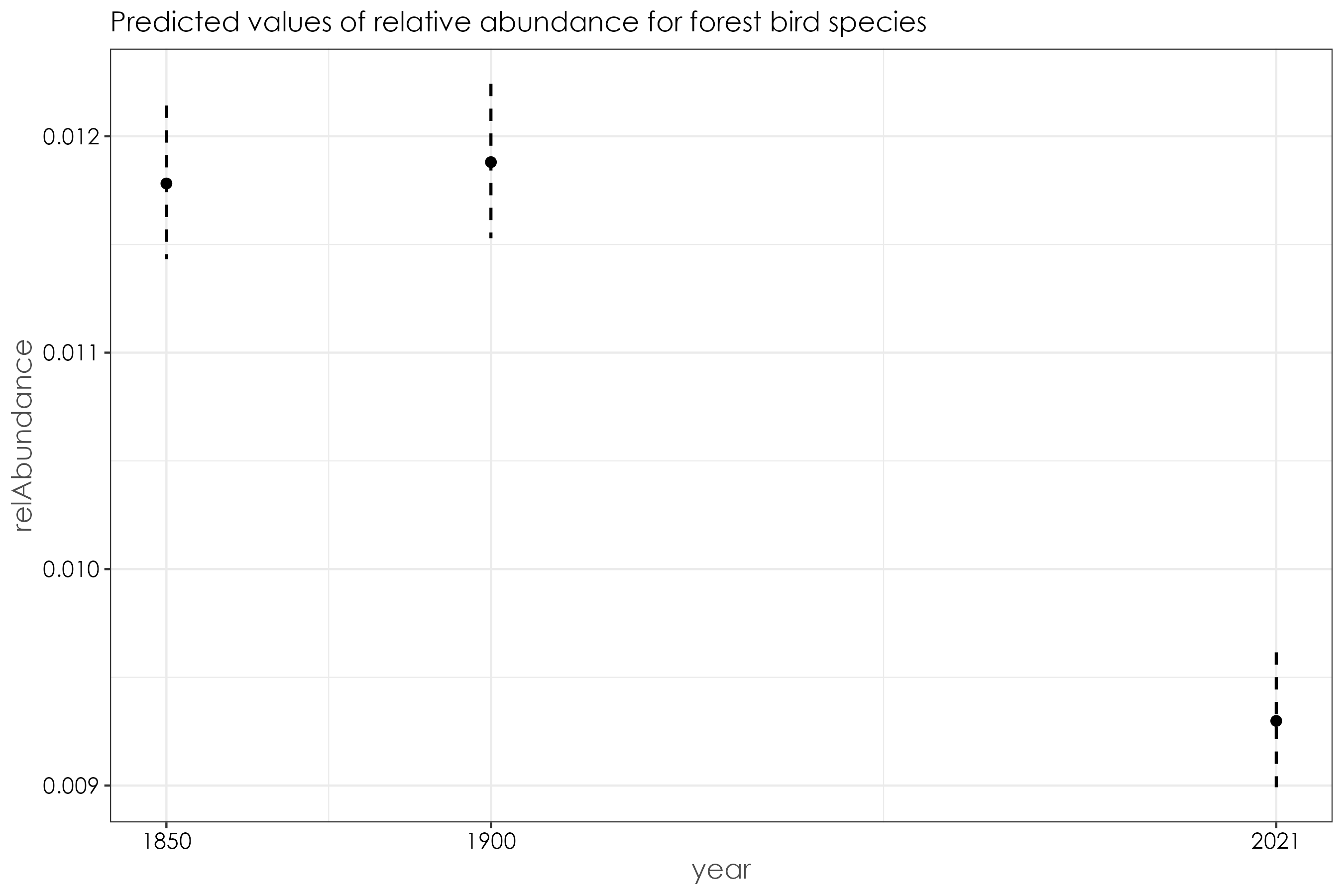
12.10 Grassland birds
grassland_birds$year <- factor(grassland_birds$year)
beta_grassland <- betareg(relAbundance ~ year, data = grassland_birds)
summary(beta_grassland)
# Call:
# betareg(formula = relAbundance ~ year, data = grassland_birds)
#
# Standardized weighted residuals 2:
# Min 1Q Median 3Q Max
# -4.6046 -0.3444 0.0742 0.1940 5.0030
#
# Coefficients (mean model with logit link):
# Estimate Std. Error z value Pr(>|z|)
# (Intercept) -4.512997 0.026442 -170.673 <2e-16 ***
# year1900 0.005862 0.036941 0.159 0.874
# year2021 -0.616943 0.043370 -14.225 <2e-16 ***
#
# Phi coefficients (precision model with identity link):
# Estimate Std. Error z value Pr(>|z|)
# (phi) 709.31 43.79 16.2 <2e-16 ***
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#
# Type of estimator: ML (maximum likelihood)
# Log-likelihood: 2316 on 4 Df
# Pseudo R-squared: 0.4275
# Number of iterations: 69 (BFGS) + 2 (Fisher scoring)
# plot predicted model estimates
fig_beta_grassland <- ggpredict(beta_grassland) %>%
plot() +
set_theme(
base = theme_bw(),
theme.font = "Century Gothic",
axis.title.size = 1.2,
axis.textsize = 1,
axis.textcolor = "black"
) +
ggtitle("Predicted values of relative abundance for grassland bird species")
ggsave(fig_beta_grassland, filename = "figs/fig_beta_relAbundance_grasslandBirds.png", width = 9, height = 6, device = png(), units = "in", dpi = 300)
dev.off()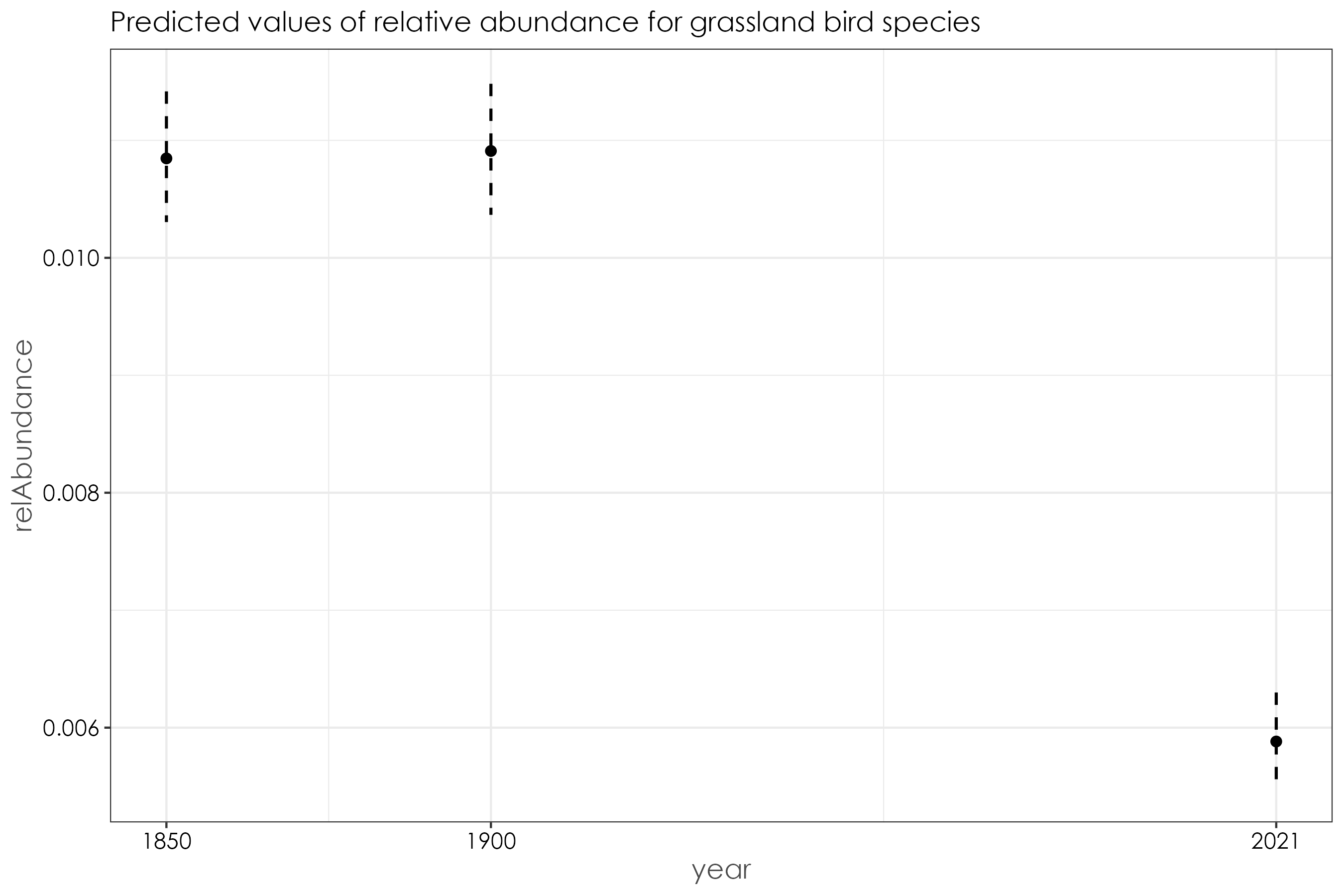
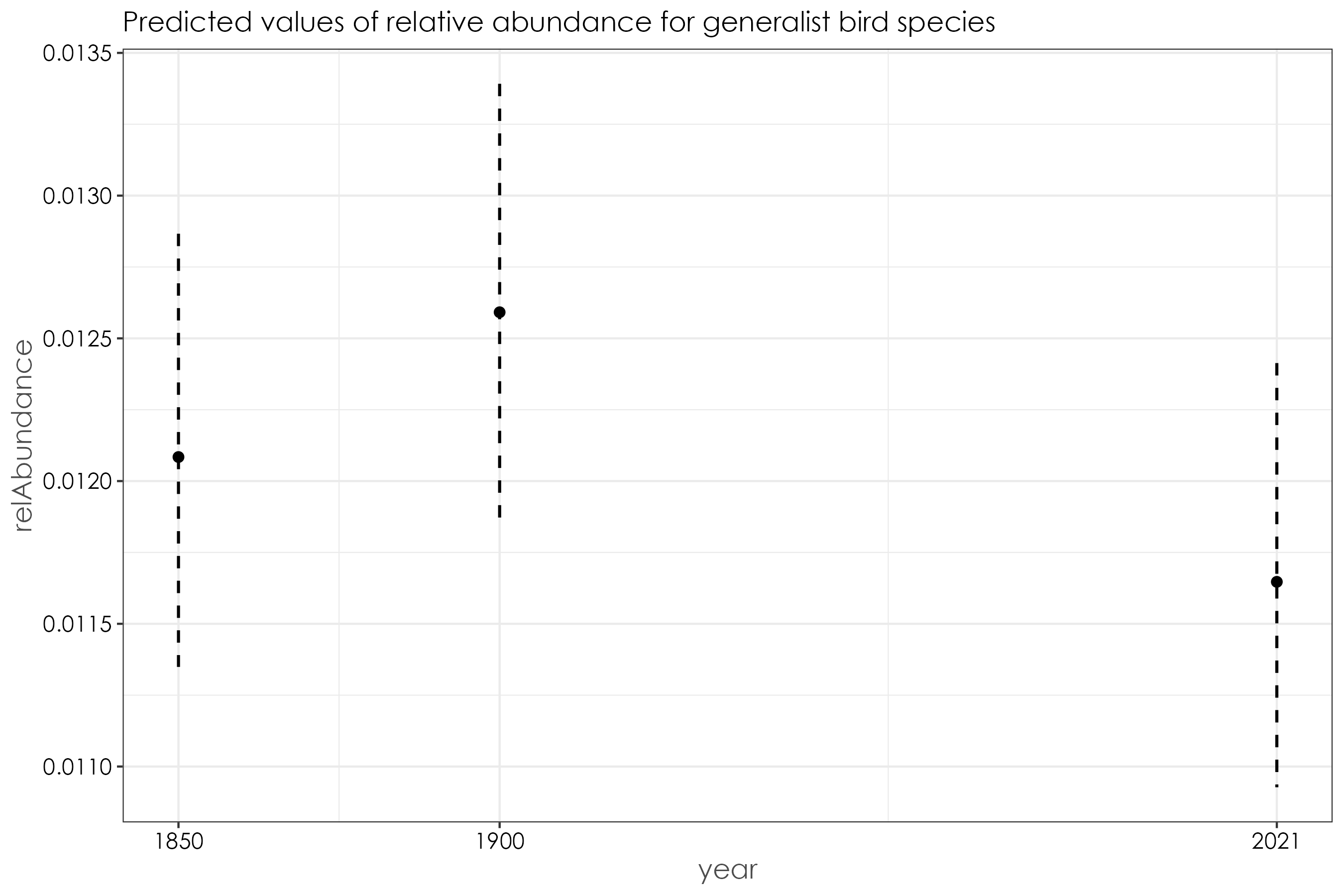
12.11 Generalist birds
generalist_birds$year <- factor(generalist_birds$year)
beta_generalist <- betareg(relAbundance ~ year, data = generalist_birds)
summary(beta_generalist)
# Call:
# betareg(formula = relAbundance ~ year, data = generalist_birds)
#
# Standardized weighted residuals 2:
# Min 1Q Median 3Q Max
# -2.1571 -0.3896 0.0175 0.1774 3.8070
#
# Coefficients (mean model with logit link):
# Estimate Std. Error z value Pr(>|z|)
# (Intercept) -4.40370 0.03241 -135.881 <2e-16 ***
# year1900 0.04163 0.04368 0.953 0.341
# year2021 -0.03730 0.04433 -0.841 0.400
#
# Phi coefficients (precision model with identity link):
# Estimate Std. Error z value Pr(>|z|)
# (phi) 180.654 7.907 22.85 <2e-16 ***
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#
# Type of estimator: ML (maximum likelihood)
# Log-likelihood: 4059 on 4 Df
# Pseudo R-squared: 0.004396
# Number of iterations: 27 (BFGS) + 2 (Fisher scoring)
# plot predicted model estimates
fig_beta_generalist <- ggpredict(beta_generalist) %>%
plot() +
set_theme(
base = theme_bw(),
theme.font = "Century Gothic",
axis.title.size = 1.2,
axis.textsize = 1,
axis.textcolor = "black"
) +
ggtitle("Predicted values of relative abundance for generalist bird species")
ggsave(fig_beta_generalist, filename = "figs/fig_beta_relAbundance_generalistBirds.png", width = 9, height = 6, device = png(), units = "in", dpi = 300)
dev.off()